Computational Research Engine (CORE™)
CORE Active Learning
CORE powers PEDAL to efficiently screen a large patient-specific
drug response space.
Helomics is now Predictive Oncology.
One of the key benefits of PEDAL is the ability to efficiently screen hundreds of compounds against thousands of patient tumor cell lines. Exploring this large, diverse drug-patient space with a traditional approach would be time-consuming and cost-prohibitive. PEDAL uses a patented Active Learning approach called CORE, developed at the Ray and Stephanie Lane Center for Computational Biology at Carnegie Mellon University, to construct predictive models of patient-specific drug response and then uses these to efficiently drive multiple rounds of drug response testing. In short, the active-learning interactively guides which wet-lab experience to do that will be the most informative to improving the overall predictive model of patient-specific drug responses that is the output of PEDAL.
CORE (Computational Research Engine) is a comprehensive in silico platform that iteratively optimizes predictive models using guided selection of experiments.
- Leverages Helomics proprietary data, collaborator and historical experimental results to produce enriched data sets.
- Learns highly accurate predictive models using a full suite of modern machine learning methods including deep learning.
- Directs compound and assay prioritization, guides experiments, concurrent optimization of properties.
- Enables informed strategy assessments, matrix optimizations, campaign termination.
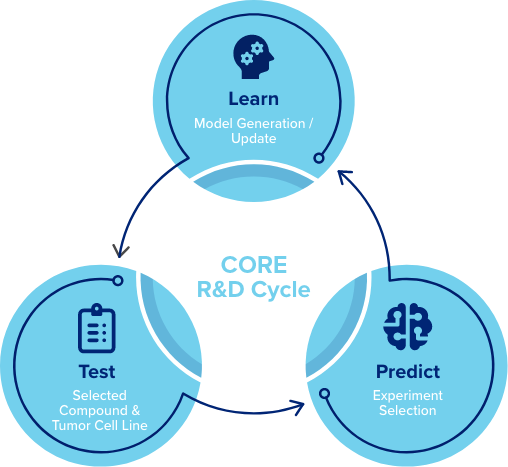
How does CORE work?
Unlike any other in silico predictive methodologies or computational methods currently in use, the active learning process driving CORE, and hence PEDAL, is iterative.
In each iteration, its algorithms recommend the most informative experiments to execute and improves the predictive model based on the results. Most current computational methods are developed only to specifically make predictions for the results of unobserved experiments, with little regard for what information might be missing. This can lead to significant errors in predictions which can have substantial costs. In contrast, CORE’s approach does not assume it has initially defined the most relevant research data. However, by directing experimentation, CORE identifies the most relevant experiments to execute— significantly improving results and accuracy while lowering costs.
- CORE employs a polypharmacological/pharmacogenomic approach which builds a large set of predictive models & selects the optimal
pairing of data and algorithm using a comprehensive machine learning
methodology. - Utilizes a diverse CORE KnowledgeBase with over 200M+ points of historical discovery data.
- Implements Active Learning to iteratively improve the predictive model for a specific campaign/application.
- Given what is already known (prior experiments + CORE KnowledgeBase) and all the experiments that could be run next, choose (iteratively) sets that are the most informative.
CORE active learning is proven in multiple projects with Pharmaceutical companies.
CORE was used in multiple projects with the Drug Safety Executive Council’s Technical Evaluation Committee, organized by Cambridge Healthtech. Studies using CORE were conducted with several members, representing three of the largest pharma companies, and yielded impressive demonstrations of CORE’s capabilities and value.
Case Study 1
Reduced Experimentation Cost to Develop Accurate Predictive Models, savings of 87%
Case Study 2
Reduced Experimentation by Leveraging Historical Experimental Results - 40% Less Experimentation Needed Using Client’s Data
Case Study 3
Reduced Compound Synthesis Required to Discover Promising Drug Leads - 30-50% Reduction in Synthesized Compounds
CoRE and Active Learning publications:
- Josh D. Kangas, Naik, Armaghan W., & Murphy, Robert. F., Efficient Discovery of Responses of Proteins to Compounds Using Active Learning, BMC Bioinformatics, 15(1), 143, May, 2014.
- Armaghan W. Naik, Kangas, Joshua D., Langmead, Christopher J. & Murphy, Robert F., Efficient Modeling and Active Learning Discovery of Biological Responses. PLoS ONE 8(12) e83996, December 2013.
- Robert F. Murphy, An Active Role for Machine Learning in Drug Development, Nature Chemical Biology, June 2011.
- J. D. Kangas, Naik, A. W. & Murphy, R. F., Active Learning to Improve Efficiency of Drug Discovery and Development, SLAS 2014 poster describing the capabilities and uses of the CoRE.
- R. J. Brennan, Kangas, J. D., Schmidt, F., Khan-Malek, R. & Keller, D. A., Applying an Active Machine Learning Process to Build Predictive Models of In Vivo Toxicity from ToxCast Screening Data, ToxCast Data Summit, September 2014. Additional Information
Start a PEDAL pilot program today
Complete the form and our PEDAL expert will be in touch to discuss customized solutions for your needs.

News & resources
“The way in which Predictive Oncology utilizes the power of predictive models offers the potential to optimize the utilization of resources, improve patient outcomes, and...
Life Sciences Knowledge Hub, recognized as the fastest-growing life sciences media platform, met with our Chief Executive Officer, Raymond F. Vennare, and Chief Business...
Our mission at Predictive Oncology is to change the landscape of oncology drug discovery and enable the development of more effective therapies for the treatment...



















