Computational Research Engine (CORE™)
CORE case studies
Learn more about our comprehensive in silico platform.
Helomics is now Predictive Oncology.
Case study 1
How does CORE work?
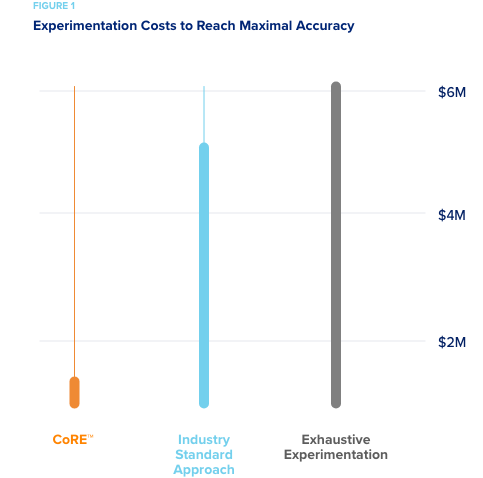
Two pharmaceutical companies wanted to compare the cost and accuracy of predictive models developed using CORE’s active machine learning methods, to those using standard industry methods. EPA’s ToxCast dataset was used as a simulated “experimental space” for the test.
- A predictive model of the experimental space was developed using the current industry standard analytic methods. Using several machine
learning approaches – RandomForest and LASSO regression– it was
necessary to explore 80% of the experimental space to reach the
maximum predictive accuracy when compounds were chosen for
experimentation based on their chemical diversity. - However, it only took 10% of the experimental space for CORE to reach this level of predictive accuracy.
Given the EPA estimate that $6M was spent on the experiments to develop ToxCast, when monetized, current industry methods would have cost $4.8M to explore and achieve the level of accuracy that CORE would have achieved for $600K. An 87% savings!
Any further experimentation directed by CORE resulted in an accuracy that was better than standard machine learning methods regardless of experiment selection methods.
Case study 2
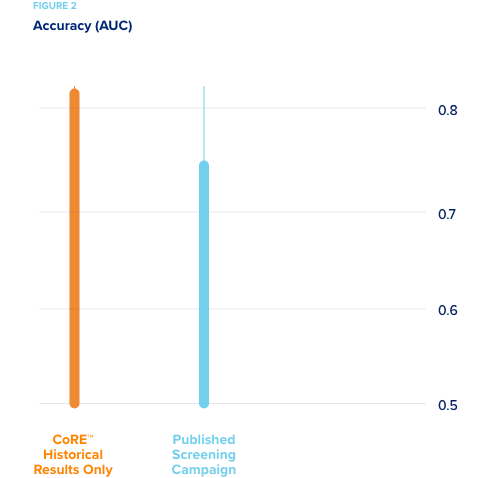
Reduced experimentation by leveraging historical experimental results.
A large pharmaceutical company wanted to compare the efficiency of CORE’s active machine learning methods to standard industry methods for predicting hepatotoxicity. Their high content screening (HCS) data from a recently published study was used.
- A predictive model of the experimental space was developed using the current industry standard analytic methods. About 50% of the
experiments executed in the Study were needed to create the most
accurate predictive model. - By comparison, it took only 30% of the experimental space for CORE to reach this same level of accuracy predicting hepatotoxicity.
While it took 40% less experimentation, the savings could not be estimated as costs were not made available. More interestingly, collaborators then suggested that methods be tested for predicting toxicity without using “new” experimental results from HCS screens. This is as if the models were developed entirely in silico without novel experimentation. In order to use only our extensive database of prior research (CORE knowledgebase) on this problem, a new, sophisticated method was designed that works with extremely sparse data sets. Using this method with the dataset and no current experimental results, CORE developed a model with higher accuracy than any methods previously tested. This shows that the knowledge gathered in their new HCS experiments was actually already in the CORE knowledgebase, but it had been gathered in different experiments, testing different compounds. The active learning methods used by CORE enabled us to capture that knowledge effectively.
Case study 3
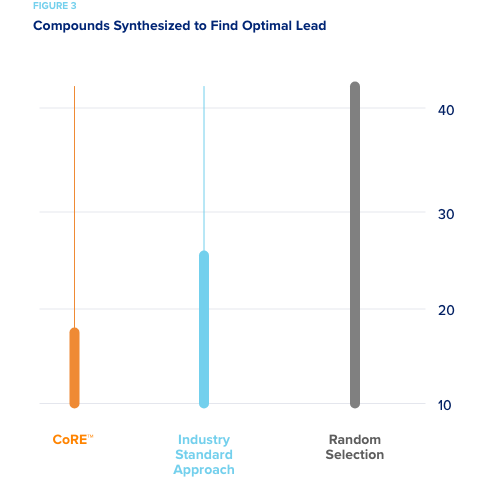
Reduced compound synthesis required to discover promising drug leads.
A smaller pharmaceutical company specializing in CNS drug development wished to assess how well CORE would have performed on a completed drug discovery campaign had it been used to direct experimentation. The pharma company conducted the campaign and identified a lead to advance after synthesizing a large number of compounds. In our simulations, CORE used their historical data to simulate an active learning approach as if it was directing compound synthesis. So all of their data was hidden from CORE and only revealed when CORE recommended a batch of compound be “synthesized.” Random selection required that on average 42 compounds be synthesized in order to predict the ideal compound. The “industry standard approach” required on average 25 compounds to be synthesized to produce an optimal lead. CORE required an average of only 18 compounds be synthesized to produce the optimal lead to advance.
This represents a 30-50% reduction in the number of synthesized compounds that would have needed to be made.
CORE and Active Learning publications:
- Josh D. Kangas, Naik, Armaghan W., & Murphy, Robert. F., Efficient Discovery of Responses of Proteins to Compounds Using Active Learning, BMC Bioinformatics, 15(1), 143, May, 2014.
- Armaghan W. Naik, Kangas, Joshua D., Langmead, Christopher J. & Murphy, Robert F., Efficient Modeling and Active Learning Discovery of Biological Responses. PLoS ONE 8(12) e83996, December 2013.
- Robert F. Murphy, An Active Role for Machine Learning in Drug Development, Nature Chemical Biology, June 2011.
- J. D. Kangas, Naik, A. W. & Murphy, R. F., Active Learning to Improve Efficiency of Drug Discovery and Development, SLAS 2014 poster describing the capabilities and uses of the CORE.
- R. J. Brennan, Kangas, J. D., Schmidt, F., Khan-Malek, R. & Keller, D. A., Applying an Active Machine Learning Process to Build Predictive Models of In Vivo Toxicity from ToxCast Screening Data, ToxCast Data Summit, September 2014. Additional Information
Start a PEDAL pilot program today
Complete the form and our PEDAL expert will be in touch to discuss customized solutions for your needs.

News & resources

Drug repurposing using the Predictive Oncology machine learning approach and proprietary biobank of frozen dissociated tumor cells (DTCs) is outlined in this new white paper....Learn more

Drug repurposing using the Predictive Oncology machine learning approach and proprietary biobank of frozen dissociated tumor cells (DTCs) is outlined in this new white paper....Learn more

Following the generation of successful results with the University of Michigan through the ACE program, Predictive Oncology is now actively calling for submissions for a...Learn more


















